GeneGlobe ID: SMH-3003AA | Cat. No.: 337021 | qBiomarker Somatic Mutation PCR Arrays
qBiomarker™ Somatic Mutation PCR Array Human Oncogene and Tumor Suppressor Panel (384-well format)
Product Specification
PCR plate and master mix
Target List
ABL1
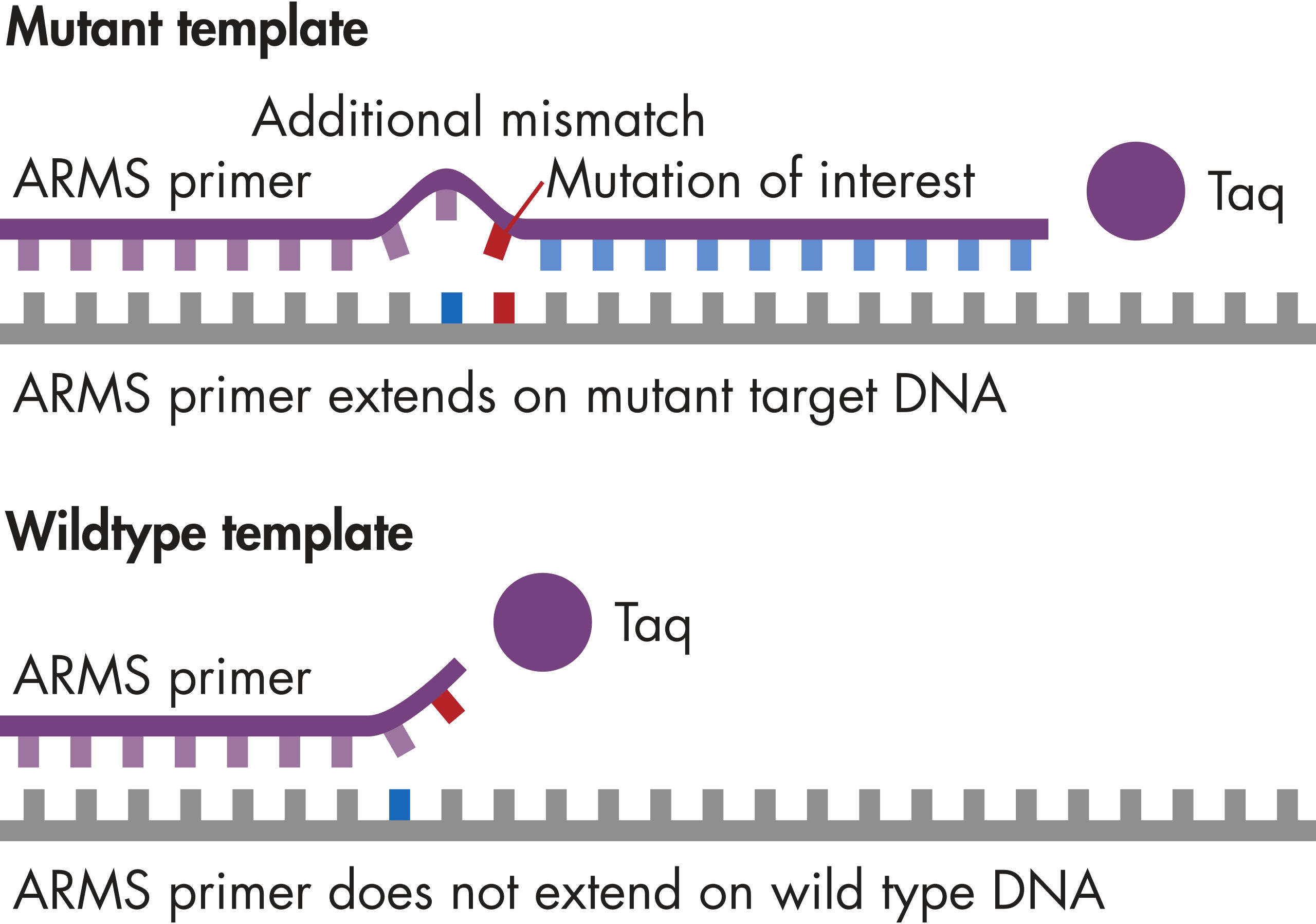
The mutations queried by these assays mostly lie in the protein kinase domain.
AKT1
The best known AKT1 mutations are c.49G>A, p.E17K, a PH domain mutation that results in constitutive targeting of AKT1 to plasma membrane, and an activating mutation, c.145G>A, p.E49K.
ALK
The most frequently observed gain-of-function ALK mutations occur in the protein’s tyrosine kinase domain from amino acids 1116 to 1383.
APC
The most commonly detected APC inactivation mutations are mainly composed of truncation mutations (due to nonsense mutations and frameshift mutations) and point mutations between codons 1250 and 1578.
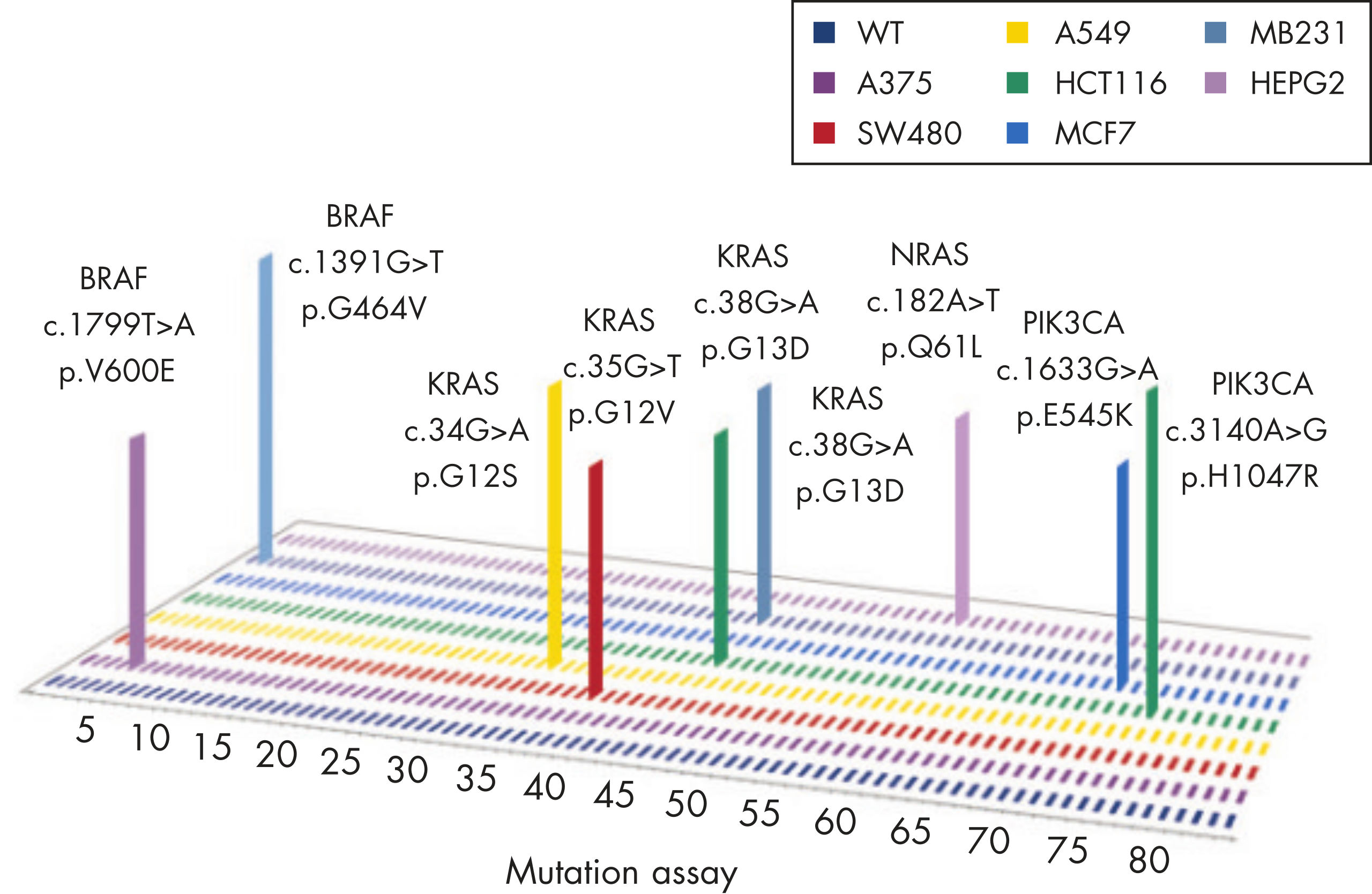
BRAF
Common point mutations in BRAF either increase kinase activity, such as L597V, L597Q and V600E, or inhibit kinase activity, such as G464V, G466V and G469A.
CDKN2A
The most important CDKN2A loss-of-function mutations occur in the consensus ankyrin domain, which lead to the inability to form stable complexes with its targets.
CSF1R
This mutation near the C-terminus that corresponds to SNP variant rs1801271 abolishes the down-regulation of activated CSF1R.
CTNNB1
The most frequently detected CTNNB1 mutations result in abnormal WNT signaling activity. The mutated codons are mainly several serines and threonines targeted for phosphorylation by GSK3B.
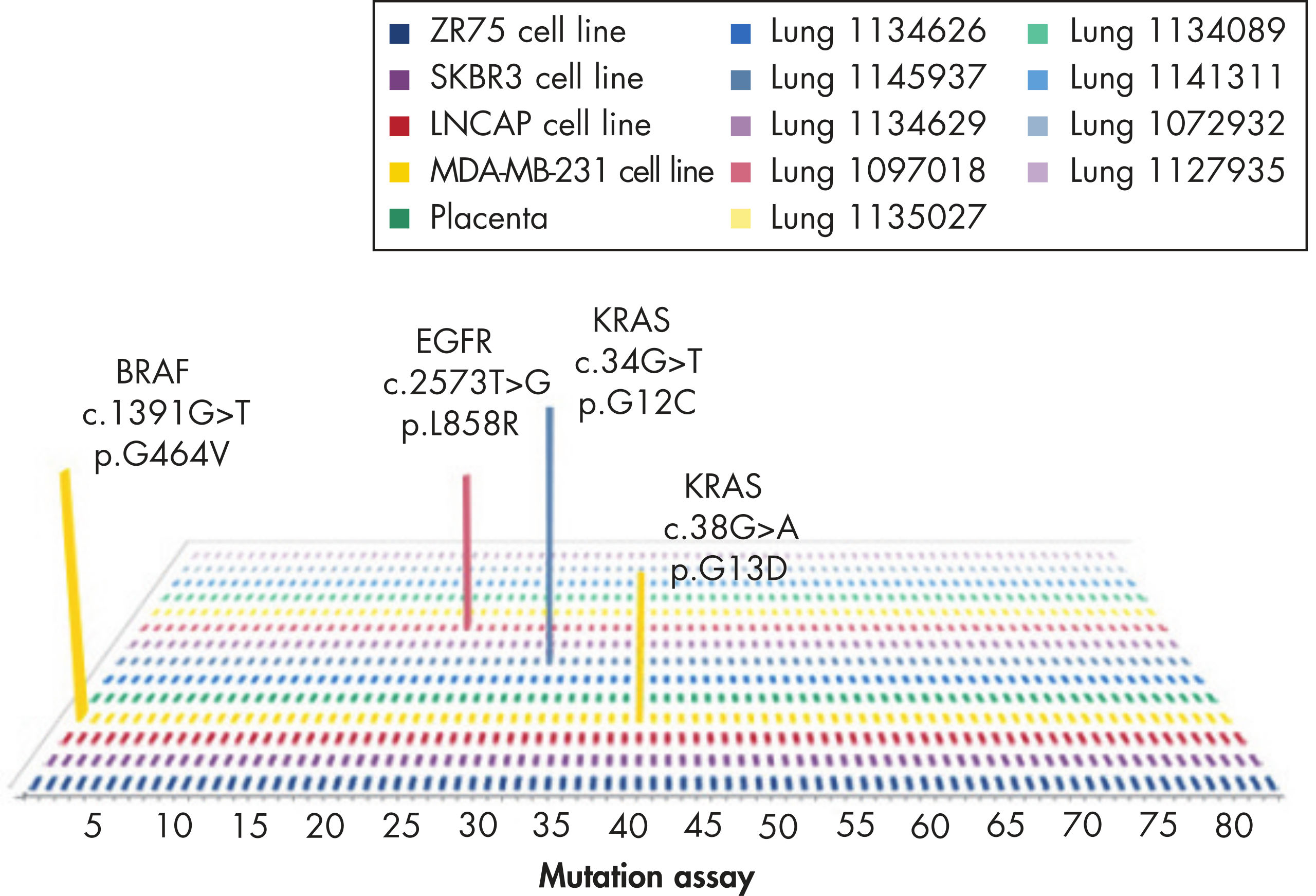
EGFR
These assays detect the most frequently identified EGFR mutations, which include P-loop and activation loop point mutations, kinase domain deletions, and insertion mutations.
ERBB2
The most frequently identified activating ERBB2 mutations cluster in its kinase domain.
EZH2
All detected mutations lie in the SET domain responsible for histone lysine methyltransferase activity.
FBXW7
These mutation assays detect FBXW7 variants in either the third or the fourth repeat of the protein's WD40 domain, which is involved in protein-protein interactions.
FGFR3
The most frequently identified FGFR mutations occur in their kinase domains and non-kinase extracellular domains such as the hinge regions and IgG-like domains. Some of the somatic mutations also correspond to congenital SNPs involved in genetic diseases.
FLT3
The most frequently identified FLT3 variants include point mutations, insertions and deletions in the juxtamembrane and activation loop domains of the protein.
GNAQ
The mutations queried by these assays all lie in the protein's GTP nucleotide binding domain.
GNAS
Mutations in this gene result in pseudohypoparathyroidism type 1a (PHP1a), which has an atypical autosomal dominant inheritance pattern requiring maternal transmission for full penetrance.
HRAS
The mutation assays detect the most important HRAS variants, all found in codons 12, 13, and 61.
IDH1
Most of these mutations abolish magnesium binding and alter the enzyme's activity to convert alpha-ketoglutarate into R(-)-2-hydroxyglutarate instead of isocitrate into alpha-ketoglutarate.
IDH2
These mutations all lie in the substrate binding domain, and one (p.R140Q) is associated with D-2-hydroxyglutaric aciduria.
JAK2
Most of these mutations lie in protein kinase domain 1. One mutation (p.V615F) confers cytokine-independent growth to BaF3 pro-B cells. Mutations at R683 lead to constitutive tyrosine phosphorylation activity promoting cytokine hypersensitivity and are associated with susceptibility to Budd-Chiari syndrome.
KIT
The most frequently identified KIT gain-of-function mutations include the D816V point mutation; point mutations in, and a deletion of, exon 11 (the juxtamembrane domain); an exon 9 insertion; as well as exon 13 point mutations.
KRAS
The mutation assays detect the most frequently occurring KRAS mutations in codons 12, 13, and 61. Mutations at these positions reduce the protein’s intrinsic GTPase activity and/or cause it to become unresponsive to RasGAP.
MET
The most frequently identified MET gain-of-function point mutations lie in its tyrosine kinase and juxtamembrane domains.
NF2
NF2 is similar to some members of the ERM family of proteins (ezrin, radixin, moesin) and links cell-surface proteins with cytoskeletal components and proteins involved in cytoskeletal dynamics. Mutations in this gene are associated with neurofibromatosis type II which is characterized by nervous system and skin tumors and ocular abnormalities.
NOTCH1
Most of the represented mutations lie in the NOTCH1 protein's predicted cytoplasmic domains.
NPM1
NPM1 encodes a phosphoprotein that shuttles between the nucleus and the cytoplasm and is thought to be involved in regulation of the ARF/p53 pathway. A number of gene fusion events with NPM1 have been characterized, in particular the anaplastic lymphoma kinase gene on chromosome 2. Mutations in this gene are associated with acute myeloid leukemia.
NRAS
The mutation assays detect the most important NRAS variants, all found in codons 12, 13, and 61.
PDGFRA
The most frequently identified PDGFRA gain-of-function variants include deletions, insertions, and point mutations in the regions of p.D842-S847 and p.R554-E571 as well as the specific point mutations p.N659Y and p.T674I.
PIK3CA
The most frequently detected PIK3CA gain-of-function mutations occur either in the kinase domain (T1025-G1049, such as H1047L and H1047R) that increase its activity or in the helical domain (P539-E545) which mimic activation by growth factors.
PTEN
The most commonly detected PTEN loss-of-function mutations either truncate the protein (such as K6fs*4, R130*, R130fs*4, R233*, P248fs*5, and V317fs*3) or cause phosphatase inactivation (such as those at R130 and R173).
RB1
The most recurrent loss-of-function mutations in the RB1 tumor suppressor gene either truncate the protein or disrupt interactions with its binding partners.
RET
The mutations p.E768D, p.A883F, and p.M918T lie in the protein kinase domain and soluble RET kinase fragment. Most of the remaining mutations lie in predicted extracellular domains.
SRC
SRC is a proto-oncogene and a tyrosine-protein kinase that plays a role in the regulation of embryonic development and cell growth. Mutations in this gene could be involved in the malignant progression of colon cancer.
STK11
The most commonly detected STK11 inactivation variants either are point mutations or truncate the protein.
TP53
The most frequently detected somatic TP53 mutations occur in the DNA-binding domain which disrupt DNA binding and/or protein structure.
VHL
Included on the panel are 6 of the most commonly detected VHL point mutations or truncation mutations that lead to loss of tumor suppressor function of the VHL protein.
Product Resources
File Size: 118.03 KBLanguage: English
Resources
Supplementary Protocols (7)
Download Files (1)
Safety Data Sheets (1)
Instrument Technical Documents (2)
Performance Data (3)
Kit Handbooks (1)
White Papers (1)
Articles (1)
Application Notes (1)
Certificates of Analysis (1)
Introducing the qBiomarker™ Somatic Mutation PCR Array Human Oncogene and Tumor Suppressor Panel (384-well format), a cutting-edge addition to our qBiomarker Somatic Mutation PCR Arrays lineup, specifically designed for Mutation/Variant Detection applications. This premium product offers unparalleled performance for researchers working with Human models. Leveraging advanced technology, the qBiomarker™ Somatic Mutation PCR Array Human Oncogene and Tumor Suppressor Panel (384-well format) facilitates accurate and reliable results, making it an indispensable tool for your scientific investigations. Whether you're conducting Mutation/Variant Detection, or any other sophisticated analyses, this product from our qBiomarker Somatic Mutation PCR Arrays collection ensures optimal efficiency and reproducibility for Human studies. Embrace the future of research with qBiomarker™ Somatic Mutation PCR Array Human Oncogene and Tumor Suppressor Panel (384-well format) and elevate your work to new heights.